Chemistry, Department of: Faculty Series
Cliff Stains Publications
ORCID IDs
Document Type
Article
Date of this Version
2006
Abstract
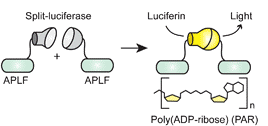
By combining custom zinc finger (ZF) DNA-binding technology [1,2] with protein fragment complementation [3], we have developed a technology, designated SEER (Sequence-Enabled Reassembly), that has the potential to “see” or detect genetic information within a living cell (Figure 1). These agents consist of two inactive parts of signal-generating peptides that have the ability to recognize specific DNA sequences. The two parts bind near each other in the presence of a user-defined DNA target site and generate a fluorescent signal. Two prototype SEER systems have been constructed, based on the reassembly of green fluorescent protein (SEERGFP) [4] and the enzyme β-lactamase (SEER-LAC). To our knowledge, these are the first examples of DNA-dependent reassembly of peptide fragments.


Comments
Published in Understanding Biology Using Peptides: American Peptide Symposia, 2006, Sylvie E. Blondelle, editor, Volume 9, Part 3, pp. 214–215; doi: 10.1007/978-0-387-26575-9_91 Copyright © 2006 Springer Verlag. Used by permission.