Chemistry, Department of: Faculty Series
David Berkowitz Publications
Document Type
Article
Date of this Version
April 2006
Abstract
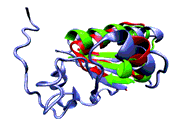
Protein structure similarity clustering (PSSC) [1]– [3] is one of a number of potential guiding principles [4], [5] that have been introduced to focus combinatorial-library design/ protein targeting. PSSC clusters protein targets with similar ligand-binding cores in which little sequence or functional similarity is evident. Lead compounds for one member of the cluster then provide novel starting points in chemical space for ligand development for other members of the PSSC. We describe herein a new clustering procedure that lends itself to ligand docking, molecular dynamics (MD), and the vector-alignment-search-tool (VAST) [6] algorithm. This MD-assisted approach offers an alternative to the serial structure retrieval/ inspection steps used to generate the original protein structure similarity cluster, centered on Cdc25A. [2] Furthermore, in a particularly difficult de novo test, the mannose 6- phosphate/insulin-like growth factor II receptor (M6P/IGF2R), lends itself to clustering, with MD opening up connections to partners that the static crystal structure coordinates fail to find.
This version includes 15 pages of Supporting Information not published with the printed edition of the article in Angewandte Chemie.


Comments
Published in Angewandte Chemie International Edition 45:46 (2006), pp. 7766–7770. Published online October 31, 2006. DOI: 10.1002/anie.200602125 Copyright © 2006 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. Used by permission.