Chemistry, Department of: Faculty Series
Robert Powers Publications
Document Type
Article
Date of this Version
2009
Citation
Proteins. 2009 April ; 75(1): 12–27. doi:10.1002/prot.22217.
Abstract
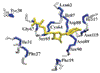
Proline utilization A (PutA) is a membrane associated multifunctional enzyme that catalyzes the oxidation of proline to glutamate in a two step process. In certain Gram-negative bacteria such as Pseudomonas putida, PutA also acts as an auto repressor in the cytoplasm when an insufficient concentration of proline is available. Here the N-terminal residues 1–45 of PutA from P. putida (PpPutA45), are shown to be responsible for DNA binding and dimerization. The solution structure of PpPutA45 was determined using NMR methods, where the protein is shown to be a symmetrical homodimer (12 kDa) consisting of two ribbon-helix-helix (RHH) structures. DNA sequence recognition by PpPutA45 was determined using DNA gel mobility shift assays and NMR chemical shift perturbations. PpPutA45 was shown to bind a 14 base-pair DNA oligomer (5′-GCGGTTGCACCTTT-3′). A model of the PpPutA45-DNA oligomer complex was generated using Haddock 2.1. The antiparallel β-sheet that results from PpPutA45 dimerization serves as the DNA recognition binding site by inserting into the DNA major groove. The dimeric core of four α-helices provides a structural scaffold for the β-sheet from which residues Thr5, Gly7, and Lys9 make sequence specific contacts with the DNA. The structural model implies flexibility of Lys9 which can either make hydrogen bond contacts with guanine or thymine. The high sequence and structure conservation of the PutA RHH domain suggest interdomain interactions play an important role in the evolution of the protein.


Comments
Copyright (c) 2008 Wiley-Liss, Inc.